Defect Finder#
As you introduce a defect into a crystalline system, all of the atoms in the system will be subject to displacements
during the atomic relaxation process. This can be especially problematic for native point defects, since the similar atoms in the simulation cell are technically fungible so split-vacancies and split-interstitials can be created with ill-defined positions.
The DefectSiteFinder can be used to identify the location of the defect in a simulation cell without any prior knowledge of how the defect is created.
This is especially useful during database building since the location of the defect is now provenance agnostic which means that the database requires less context information during the build process.
To use the DefectSiteFinder, simply instantiate the class and call the find_defect_fpos method with the defective and pristine structures as arguments. We show that the DefectSiteFinder can be used to identify the location of nitrogen vacancies in wz-GaN structure blow.
from pathlib import Path
import numpy as np
TEST_FILES = Path("../../../tests/test_files")
from pymatgen.analysis.defects.finder import DefectSiteFinder
from pymatgen.core.structure import Structure
base_struct = Structure.from_file(TEST_FILES / "v_N_GaN/bulk/CONTCAR.gz")
finder = DefectSiteFinder()
for q in range(-1, 3):
fname = f"{TEST_FILES!s}/v_N_GaN/q={q}/CONTCAR.gz"
struct = Structure.from_file(fname)
fpos = finder.get_defect_fpos(defect_structure=struct, base_structure=base_struct)
fpos -= np.round(fpos)
print(fpos)
---------------------------------------------------------------------------
ImportError Traceback (most recent call last)
Cell In[1], line 11
8 from pymatgen.core.structure import Structure
10 base_struct = Structure.from_file(TEST_FILES / "v_N_GaN/bulk/CONTCAR.gz")
---> 11 finder = DefectSiteFinder()
13 for q in range(-1, 3):
14 fname = f"{TEST_FILES!s}/v_N_GaN/q={q}/CONTCAR.gz"
File ~/work/pymatgen-analysis-defects/pymatgen-analysis-defects/pymatgen/analysis/defects/finder.py:61, in DefectSiteFinder.__init__(self, symprec, angle_tolerance)
59 if SOAP is None:
60 msg = "dscribe is required to use DefectSiteFinder. Install with ``pip install dscribe``."
---> 61 raise ImportError(
62 msg,
63 )
64 self.symprec = symprec
65 self.angle_tolerance = angle_tolerance
ImportError: dscribe is required to use DefectSiteFinder. Install with ``pip install dscribe``.
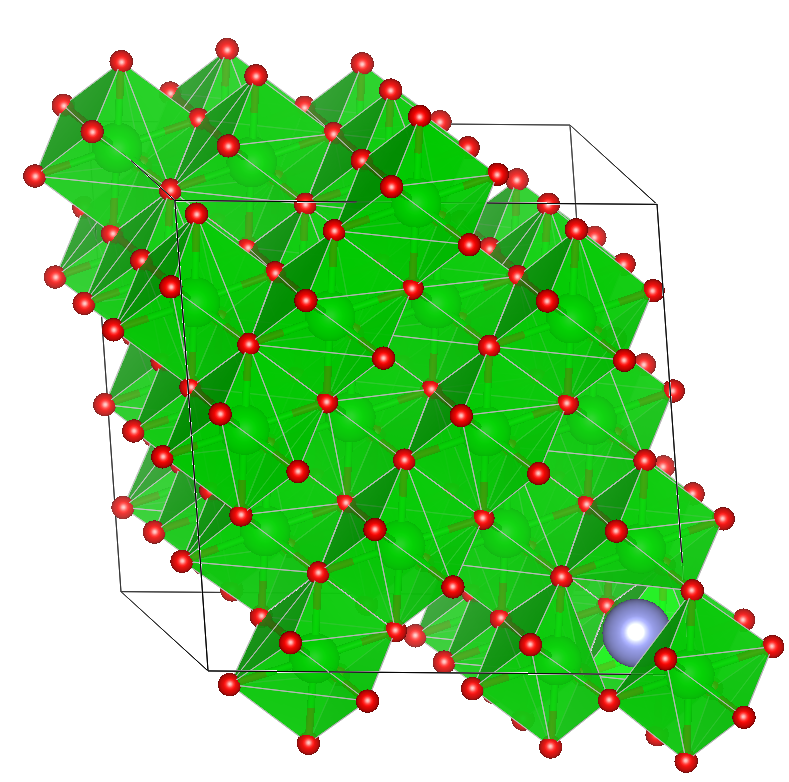
The last structure (for q=2) demonstrates that doing this in a smaller simulation cell with large distortions can be problematic so always double-check your results.
On a larger ZrO2 cell, the identified vacancy is shown
How does it work?#
The steps of the structure analysis and comparison procedure are as follows:
Calculate the anonymized SOAP vector for each symmetry distinct site in the pristine structure.
Calculate the SOAP vector for each site in the defective structure.
For each site in the defective structure, find the closest match in the pristine structure using the cosine similarity of the SOAP vectors. This cosine similarity becomes a measure of how defective the site is (how different from a bulk site).
Rank the sites by their defectiveness as defined above.
Compute the periodic average of a subset of the most defective sites.
So in a sense, we are using SOAP to arrive at a distortion field for the defect structure and calculating the center of that distortion field. This is a fairly straightforward approach, but it seems to work well in practice.